Instructions
Summary
This application allows the user to search the catalog of hemibrain body IDs and return MCFO match information in two forms:- PDF files containing match imagery, ranks, and scores
- Excel spreadsheets containing stock name, slide code, and rank
Data
We provide PatchPerPixMatch search results for ~30,000 target neuron morphologies from the hemibrain [1] version 1.2 (available at https://neuprint.janelia.org) vs ~20,000 MCFO brain acquisitions of sparse and medium dense GAL4 lines (Gen1 MCFO Phase 1 Categories 2 and 3 as published with [2], available at https://gen1mcfo.janelia.org, registered to the JRC2018 Unisex template [3]).For each of ~30,000 hemibrain body IDs, there is a PDF and a spreadsheet (xlsx) showing the best matching MCFO samples.
Content of the PDFs
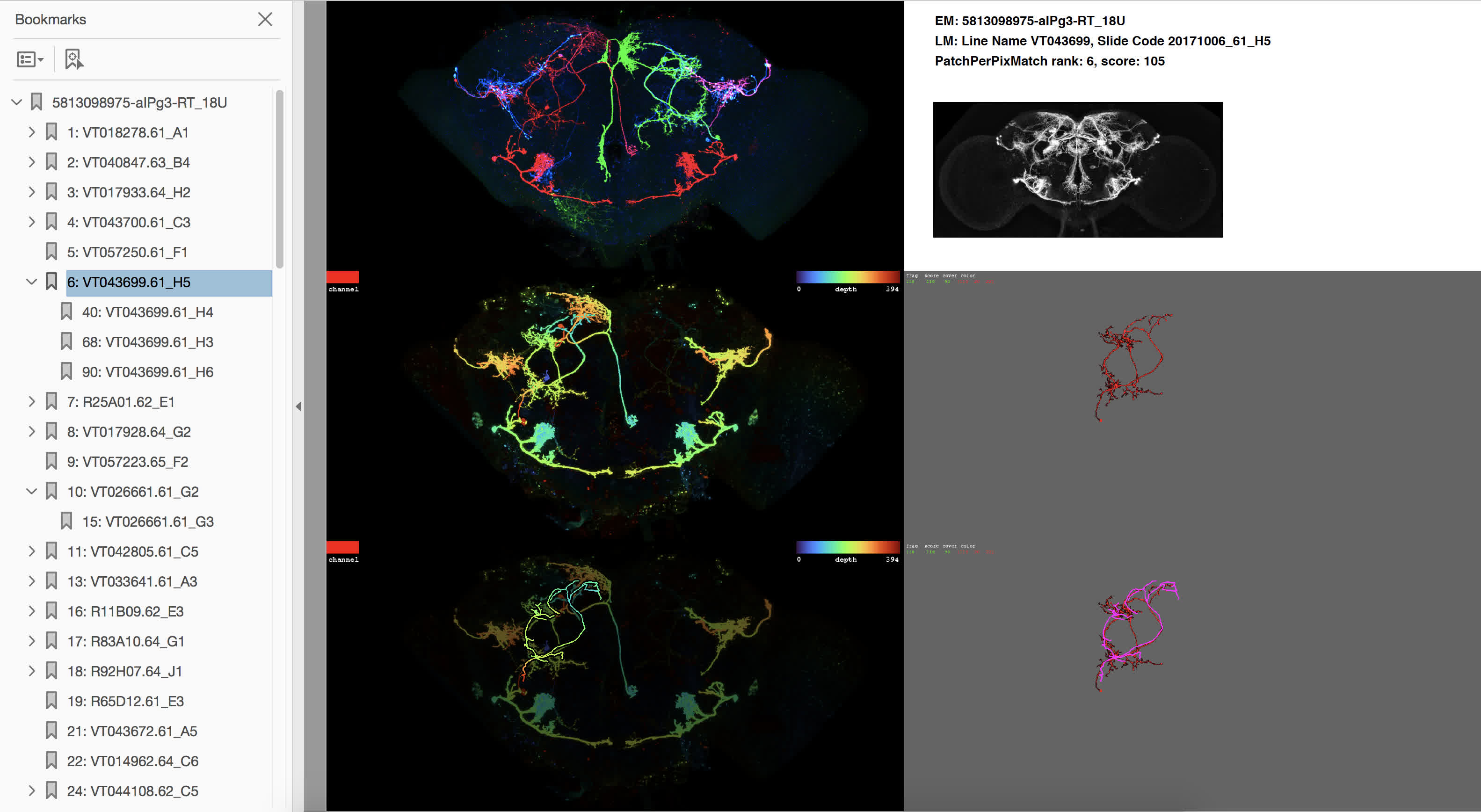
- Top left image: Maximum intensity projection (MIP) of the MCFO sample (all three neuron signal channels).
- Mid left image: Color-depth MIP of best-matching channel (i.e. projection image with depth encoded as color, as employed in [4]). The channel color is indicated in the top left corner of the image. Color depth encoding is specified in the top right corner of the image.
- Bottom left image: Color MIP of best-matching channel, dimmed and overlaid with color-depth projection of target neuron morphology.
- Top right panel: hemibrain ID of the target neuron, line name and sample ID of the MCFO sample, PatchPerPixMatch rank and score, as well as a MIP of the full expression pattern of the GAL4 line (to get a sense of the respective density).
- Mid right image: MIP of the MCFO sample masked by best-matching PatchPerPix [5] fragments.
- Bottom right image: MIP of the MCFO sample masked by best-matching PatchPerPix fragments (sometimes pruned), overlaid with target neuron morphology in purple.
Content of the spreadsheets
Spreadsheets are provided to facilitate annotation efforts. Each row contains line name, slide code, and PatchPerPixMatch rank of a match as contained in the PDF, sorted by rank.Which hemibrain body IDs are included
A list of ~30,000 hemibrain version 1.2 body IDs selected from neuprint (https://neuprint.janelia.org) by Hideo Otsuna by means of neuprint neuron quality tags, size, etc. For the same list of body IDs, Color depth MIP mask search results are published on NeuronBridge (https://neuronbridge.janelia.org).Which MCFO samples are included
~20,000 MCFO brain samples of sparse and medium dense Generation 1 lines (Gen1 MCFO Phase 1 Categories 2 and 3 as published with [2], available at https://gen1mcfo.janelia.org).How the PatchPerPixMatch results were generated
- PatchPerPix segmentations of the MCFO samples: https://github.com/Kainmueller-Lab/PatchPerPix_experiments
- PatchPerPixMatch search results: https://github.com/Kainmueller-Lab/PatchPerPixMatch
Downloading files
After a search, a table will show body ID and neuron name, and provide links to the PDF file and spreadsheet. To download a PDF file or spreadsheet, just click on the link.If you're using Chrome and want to download files in batch, we recommend Batch Link Downloader. Once you have it installed, any time you're on a page with links, you can set up for batch download by clicking on the Batch Link Download icon in your Chrome menu
 . You'll see a pop-up menu similar to this:
. You'll see a pop-up menu similar to this:
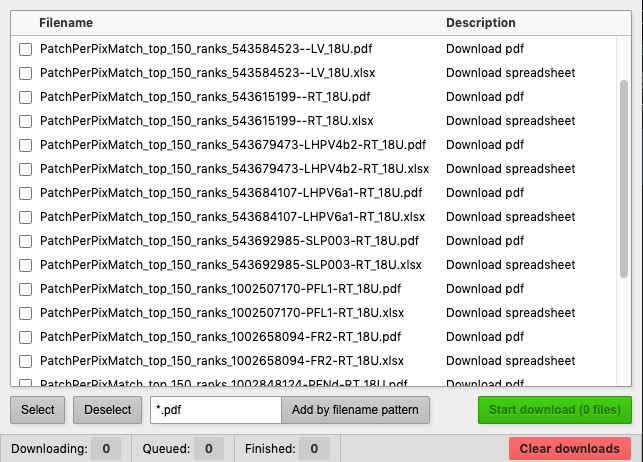
Simply check the files you want to download, then click the green "Start Download" button.
If you have any questions, please contact lisa.mais@mdc-berlin.de or kainmuellerd@janelia.hhmi.org.
If you are using PatchPerPixMatch search results in your work, please cite:
Lisa Mais, Peter Hirsch, Claire Managan, Kaiyu Wang, Konrad Rokicki, Robert R. Svirskas, Barry J. Dickson, Wyatt Korff, Gerald M. Rubin, Gudrun Ihrke, Geoffrey W. Meissner, Dagmar Kainmueller:
PatchPerPixMatch for Automated 3d Search of Neuronal Morphologies in Light Microscopy. bioRxiv. 2021: 2021.07.23.453511. DOI: 10.1101/2021.07.23.453511
References
[1] Scheffer, L. K., Xu, C. S., Januszewski, M., Lu, Z., Takemura, S., Hayworth, K. J., Huang, G. B., Shinomiya, K., Maitlin-Shepard, J., Berg, S., Clements, J., Hubbard, P. M., Katz, W. T., Umayam, L., Zhao, T., Ackerman, D., Blakely, T., Bogovic, J., Dolafi, T., Kainmueller, D., Kawase, T., Khairy, K. A., Leavitt, L., Li, P. H., Lindsey, L., Neubarth, N., Olbris, D. J., Otsuna, H., Trautman, E. T., Ito, M., Bates, A. S., Goldammer, J., Wolff, T., Svirskas, R., Schlegel, P., Neace, E., Knecht, C. J., Alvarado, C. X., Bailey, D. A., Ballinger, S., Borycz, J. A., Canino, B. S., Cheatham, N., Cook, M., Dreher, M., Duclos, O., Eubanks, B., Fairbanks, K., Finley, S., Forknall, N., Francis, A., Hopkins, G. P., Joyce, E. M., Kim, S., Kirk, N. A., Kovalyak, J., Lauchie, S. A., Lohff, A., Maldonado, C., Manley, E. A., McLin, S., Mooney, C., Ndama, M., Ogundeyi, O., Okeoma, N., Ordish, C., Padilla, N., Patrick, C. M., Paterson, T., Phillips, E. E., Phillips, E. M., Rampally, N., Ribeiro, C., Robertson, M. K., Rymer, J. T., Ryan, S. M., Sammons, M., Scott, A. K., Scott, A. L., Shinomiya, A., Smith, C., Smith, K., Smith, N. L., Sobeski, M. A., Suleiman, A., Swift, J., Takemura, S., Talebi, I., Tarnogorska, D., Tenshaw, E., Tokhi, T., Walsh, J. J., Yang, T., Horne, J. A., Li, F., Parekh, R., Rivlin, P. K., Jayaraman, V., Costa, M., Jefferis, G. S., Ito, K., Saalfeld, S., George, R., Meinertzhagen, I. A., Rubin, G. M., Hess, H. F., Jain, V. & Plaza, S. M. A connectome and analysis of the adult Drosophila central brain. eLife. 2020; 9: elife.57443. DOI: 10.7554/eLife.57443[2] Meissner, G. W., Dorman, Z., Nern, A., Forster, K., Gibney, T., Jeter, J., Johnson, L., He, Y., Lee, K., Melton, B., Yarbrough, B., Clements, J., Goina, C., Otsuna, H., Rokicki, K., Svirskas, R. R., Aso, Y., Card, G. M., Dickson, B. J., Ehrhardt, E., Goldammer, J., Ito, M., Korff, W., Minegishi, R., Namiki, S., Rubin, G. M., Sterne, G., Wolff, T., Malkesman, O. & FlyLight Project Team. An image resource of subdivided Drosophila GAL4-driver expression patterns for neuron-level searches. bioRxiv. 2020: 2020.05.29.080473. DOI: 10.1101/2020.05.29.080473
[3] Bogovic, J. A., Otsuna, H., Heinrich, L., Ito, M., Jeter, J., Meissner, G., Nern, A., Colonell, J., Malkesman, O., Ito, K. & Saalfeld, S. An unbiased template of the Drosophila brain and ventral nerve cord. PLoS ONE. 2020; 15: e0236495. DOI: 10.1371/journal.pone.0236495
[4] Otsuna, H., Ito, M. & Kawase, T. Color depth MIP mask search: a new tool to expedite Split-GAL4 creation. bioRxiv. 2018: 318006. DOI: 10.1101/318006
[5] Mais, L., Hirsch, P., Kainmueller, D.: PatchPerPix for instance segmentation. Lecture Notes in Computer Science 12370, 288–304 (2020).
